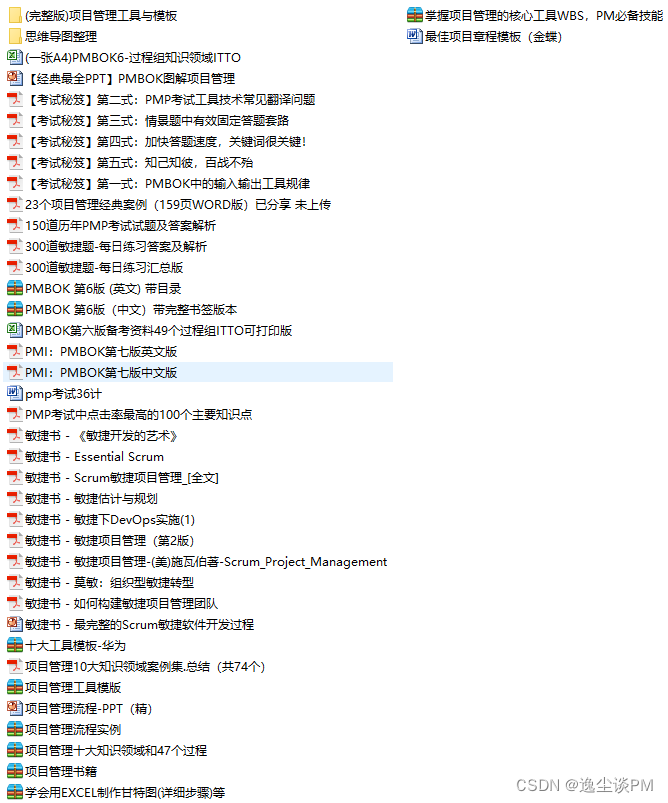
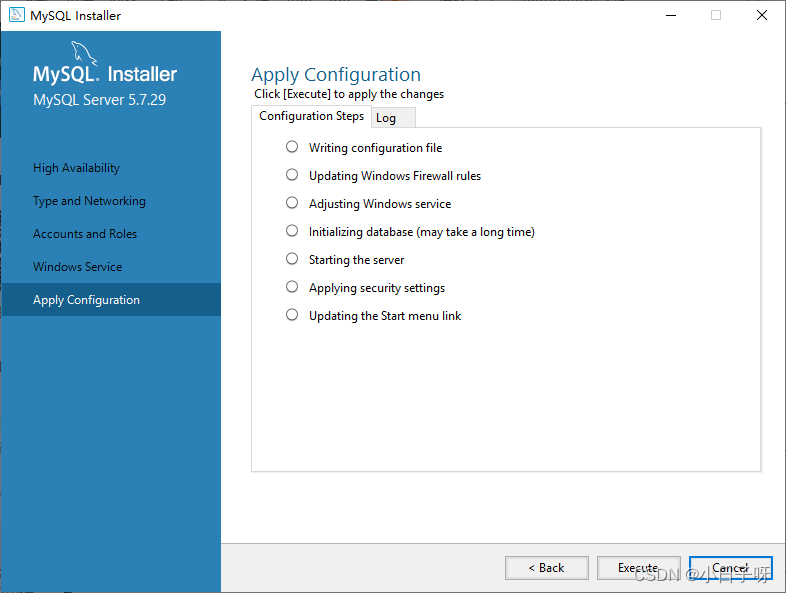
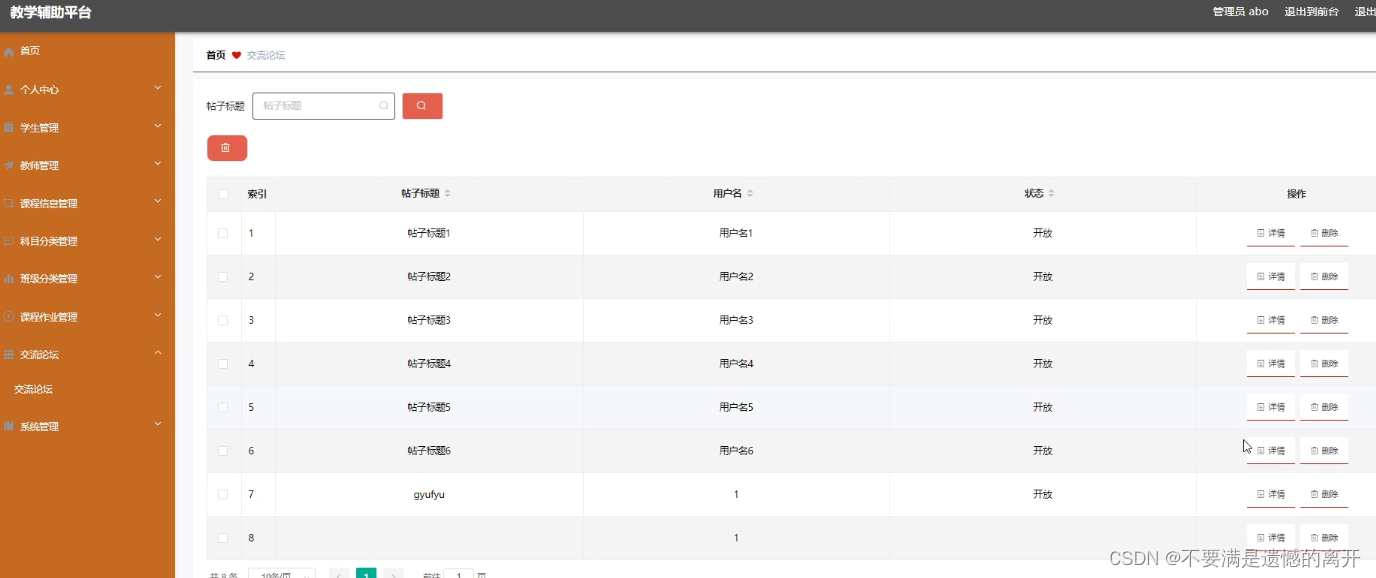
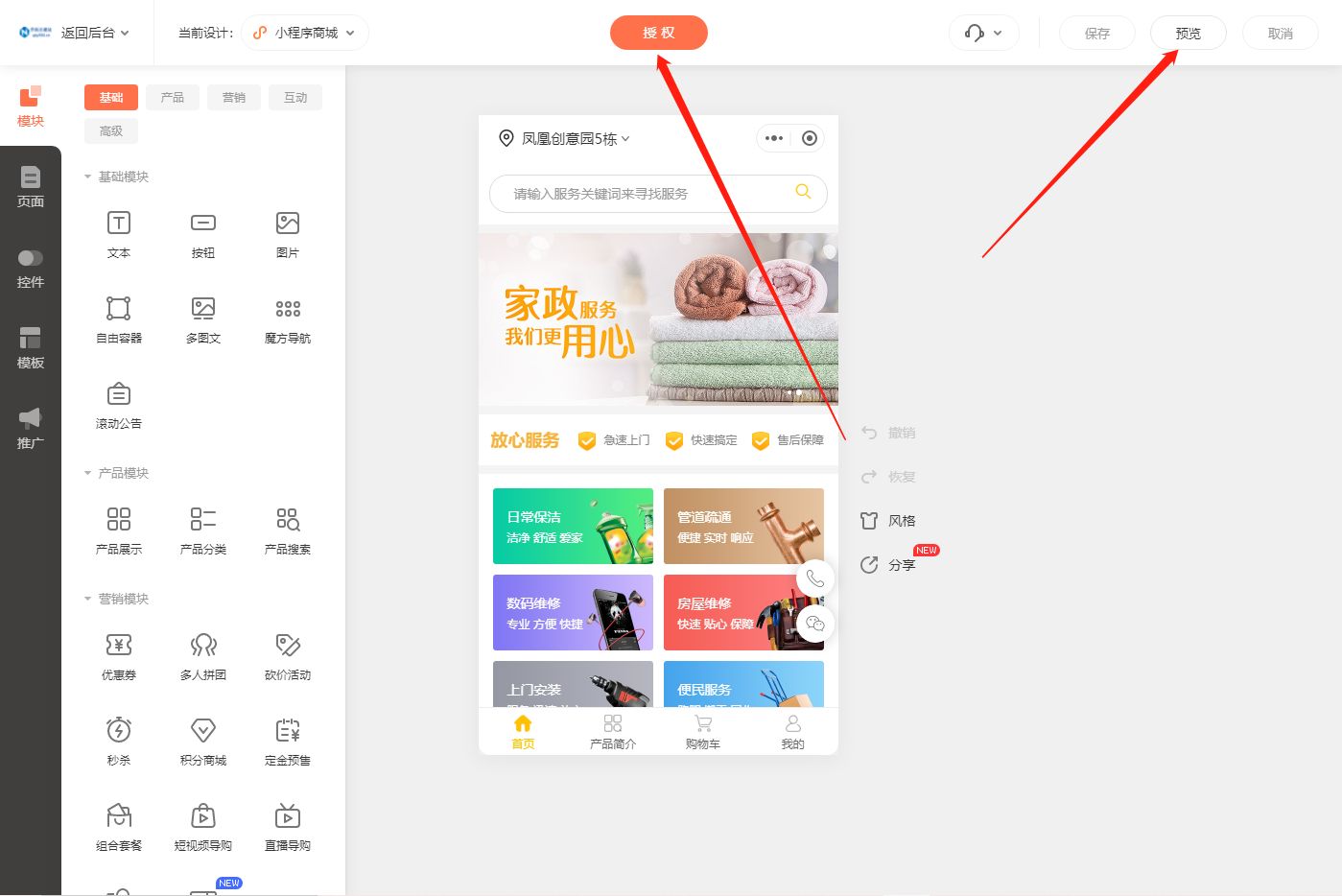
1.5 ##富集分析----------
#BiocManager::install("gprofiler2")
library(gprofiler2)
## We can perform an enrichment analyses with the genes in the complex
EnrichmentResults <- gprofiler2::gost(genes_complex, significant = TRUE,
user_threshold = 0.001, correction_method = c("fdr"),
sources=c("GO:BP","GO:CC","GO:MF"))
#1 complex------------
## We check the different complexes databases
get_complex_resources()
## We query and store complexes from some sources into a dataframe.
1.2
complexes <- import_omnipath_complexes(resources=c("CORUM", "hu.MAP"))
complexes_all <- import_omnipath_complexes(resources=get_complex_resources())
## We check all the molecular complexes where a set of genes participate
query_genes <- c("WRN","PARP1")
1.3
## Complexes where any of the input genes participate
complexes_query_genes_any <- unique(get_complex_genes(complexes,query_genes,
total_match=FALSE))
head(complexes_query_genes_any)
1.4
## Complexes where all the input genes participate jointly
complexes_query_genes_join <- unique(get_complex_genes(complexes,query_genes,
total_match=TRUE))
## We print the components of the different selected components
complexes_query_genes_join$components_genesymbols
genes_complex <-
unlist(strsplit(complexes_query_genes_join$components_genesymbols, "_"))
1.5 ##富集分析----------
#BiocManager::install("gprofiler2")
library(gprofiler2)
## We can perform an enrichment analyses with the genes in the complex
EnrichmentResults <- gprofiler2::gost(genes_complex, significant = TRUE,
user_threshold = 0.001, correction_method = c("fdr"),
sources=c("GO:BP","GO:CC","GO:MF"))
## We show the most significant results
EnrichmentResults$result %>%
dplyr::select(term_id, source, term_name,p_value) %>%
dplyr::top_n(5,-p_value)