今天为了画这个cluster中怎么显示标签的图,研究了一个Seurat中怎么画这个图的,下面是学习过程中做的总结
运行例子
rm(list=ls())
library(Seurat)
library(SeuratData)
library(ggplot2)
library(patchwork)
pbmc3k.final <- LoadData("pbmc3k", type = "pbmc3k.final")
#print(DimPlot(pbmc3k.final,label = TRUE, repel = TRUE))
# remove the legend
p=DimPlot(pbmc3k.final,label = TRUE, repel = TRUE)+theme(legend.position = "none")
print(p)
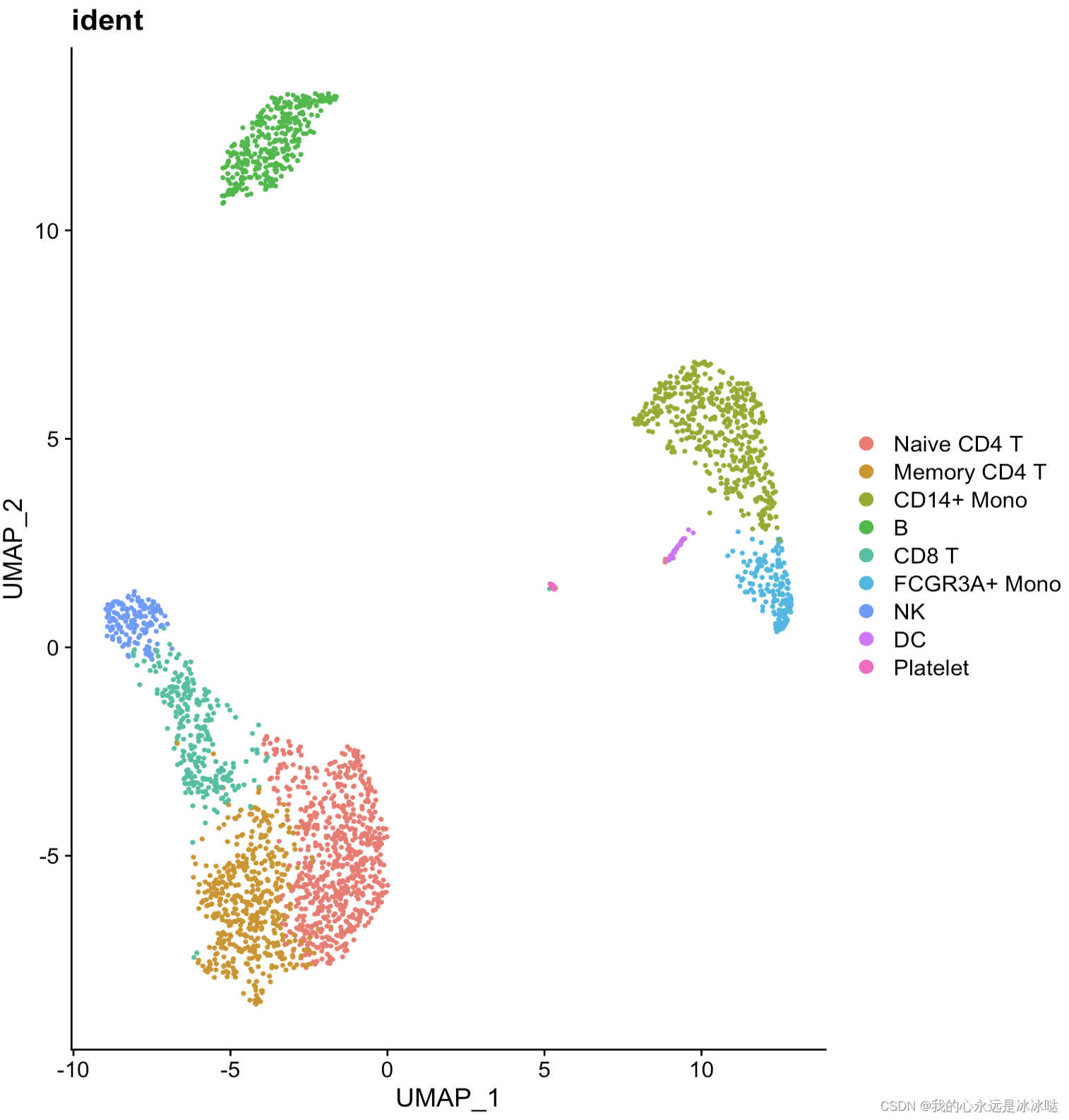
结果如下
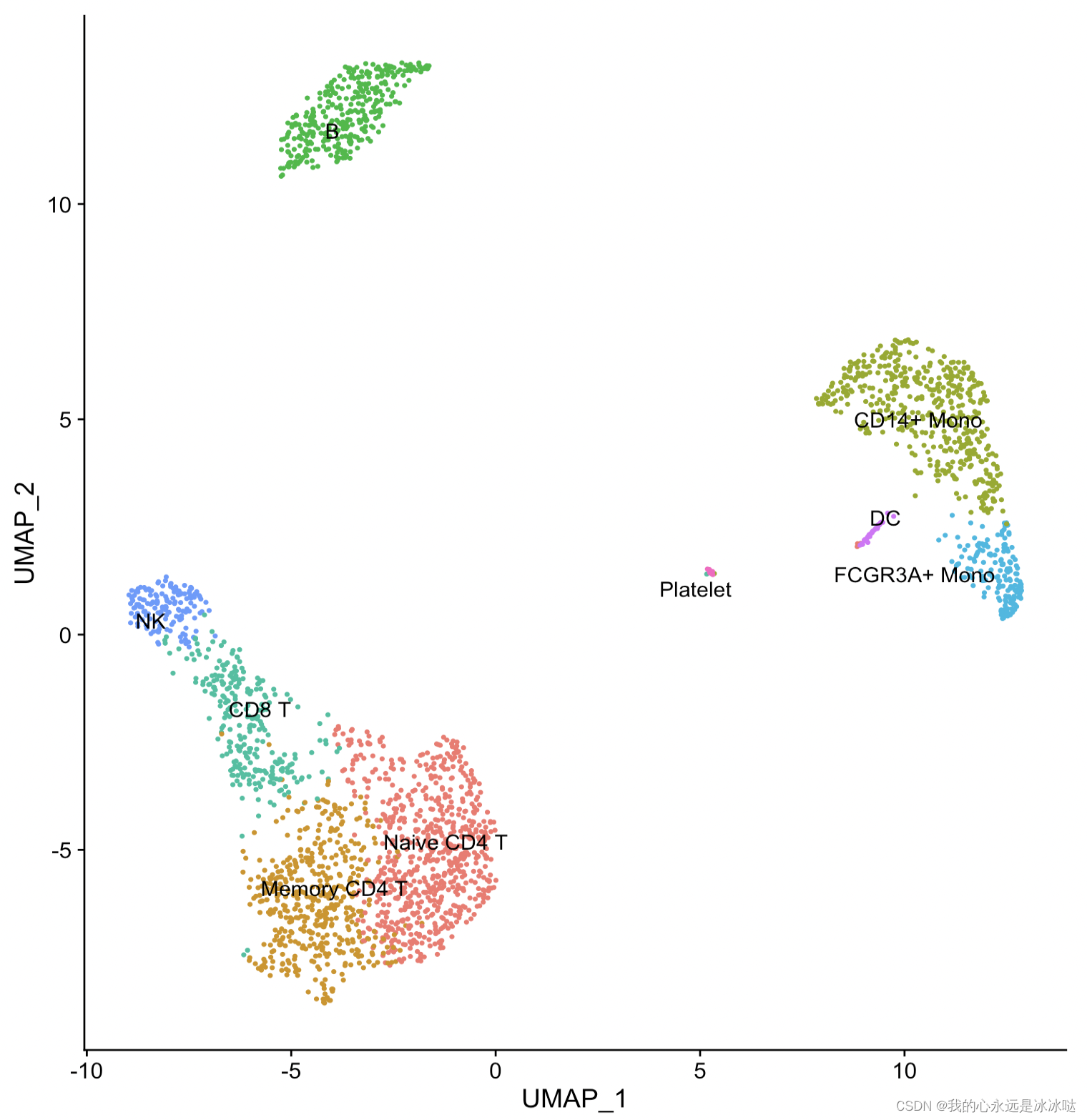 这个就是我想要达到的效果,把类别的标签放在类的中心显示,但是这个是Seurat包自己实现的,我要做的事就是把Seurat包的这个Dimplot函数给抽出来,变成自己的ggplot函数
这个就是我想要达到的效果,把类别的标签放在类的中心显示,但是这个是Seurat包自己实现的,我要做的事就是把Seurat包的这个Dimplot函数给抽出来,变成自己的ggplot函数
调试
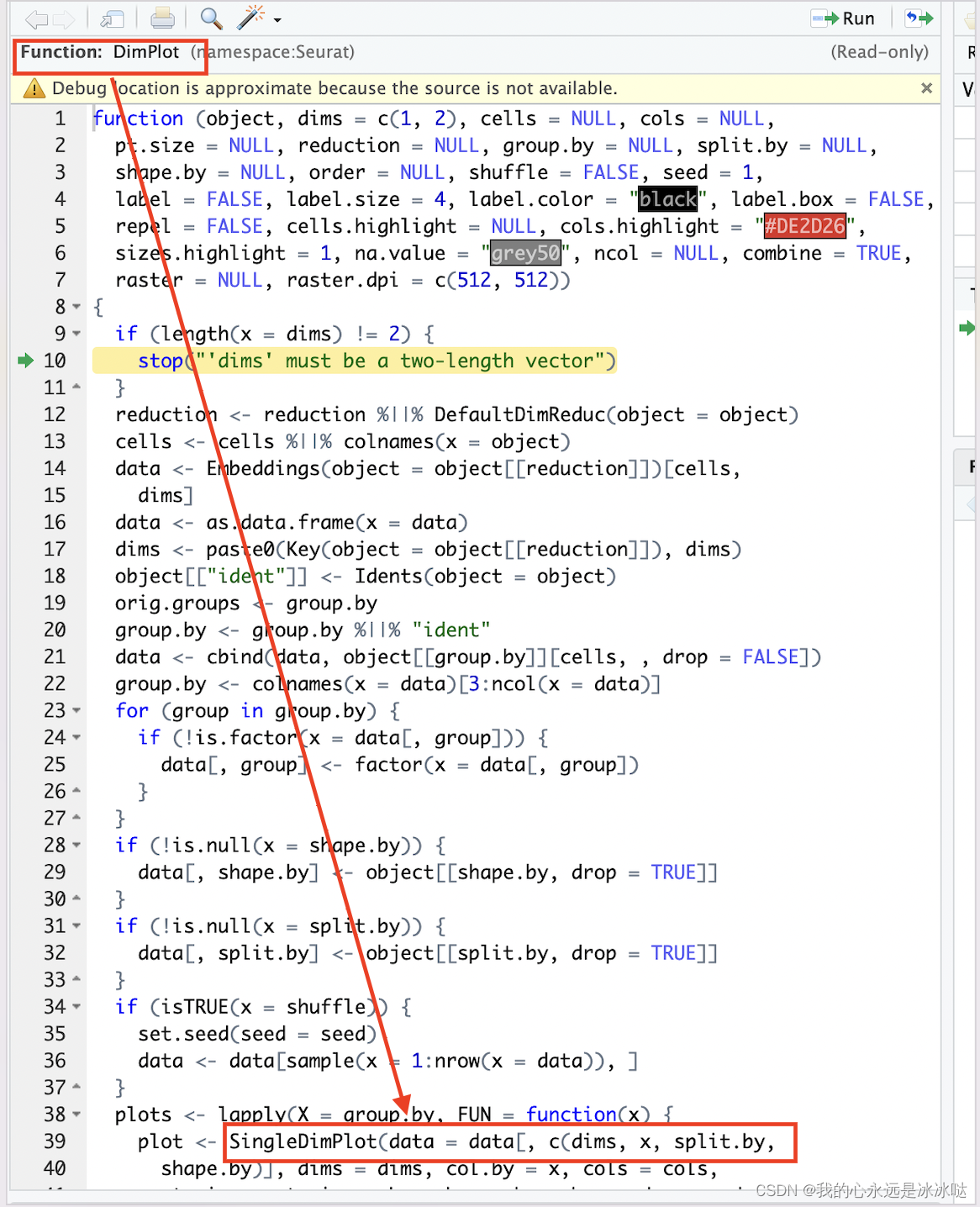 其中核心部分是下面两个函数
其中核心部分是下面两个函数
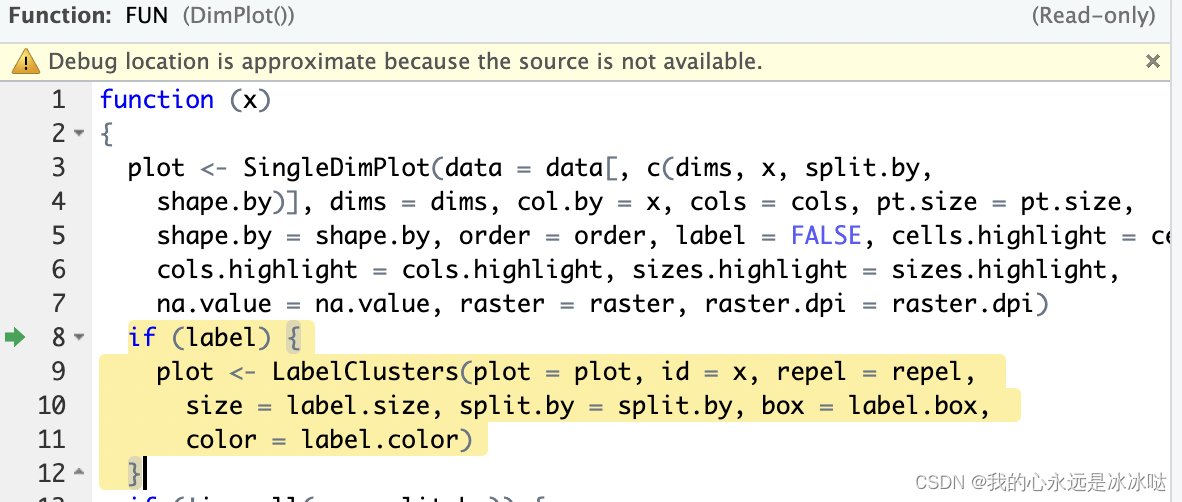 一个是
一个是SingleDimPlot函数, 另一个是LabelClusters函数,
首先发现Dimplot函数会进入函数SingleDimPlot函数,其函数实现如下
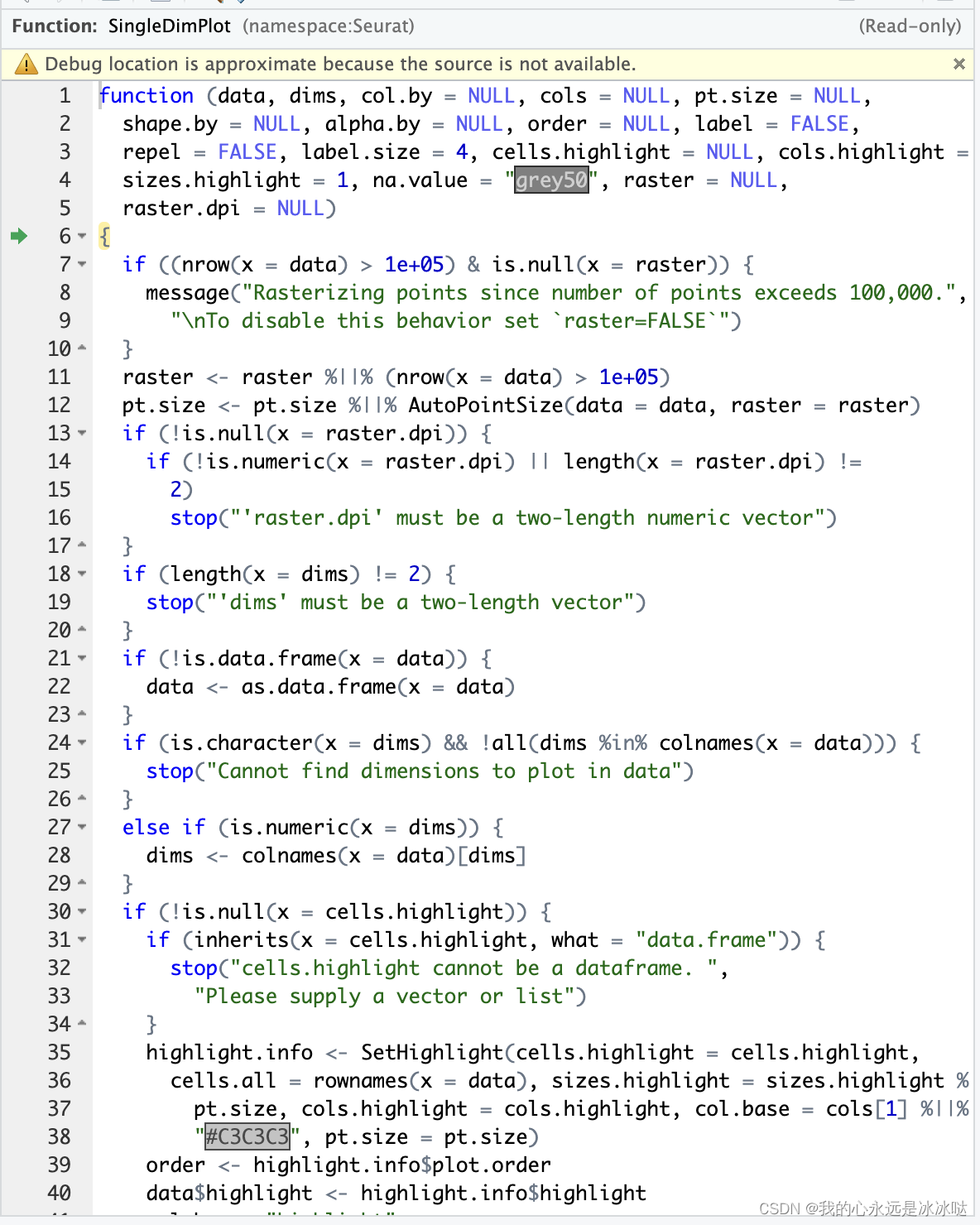 虽然这个函数看起来很复杂,其实大部分都是在判断条件,所以对于自己的需求而言,很多代码可以全删了,经过
虽然这个函数看起来很复杂,其实大部分都是在判断条件,所以对于自己的需求而言,很多代码可以全删了,经过SingleDimPlot函数后,画出来的效果如图
 可以看到,此时这个结果只是普普通通的ggplot画图,所以调整图例到细胞的类别中是在
可以看到,此时这个结果只是普普通通的ggplot画图,所以调整图例到细胞的类别中是在LabelClusters函数里, 其中LabelClusters函数实现如下
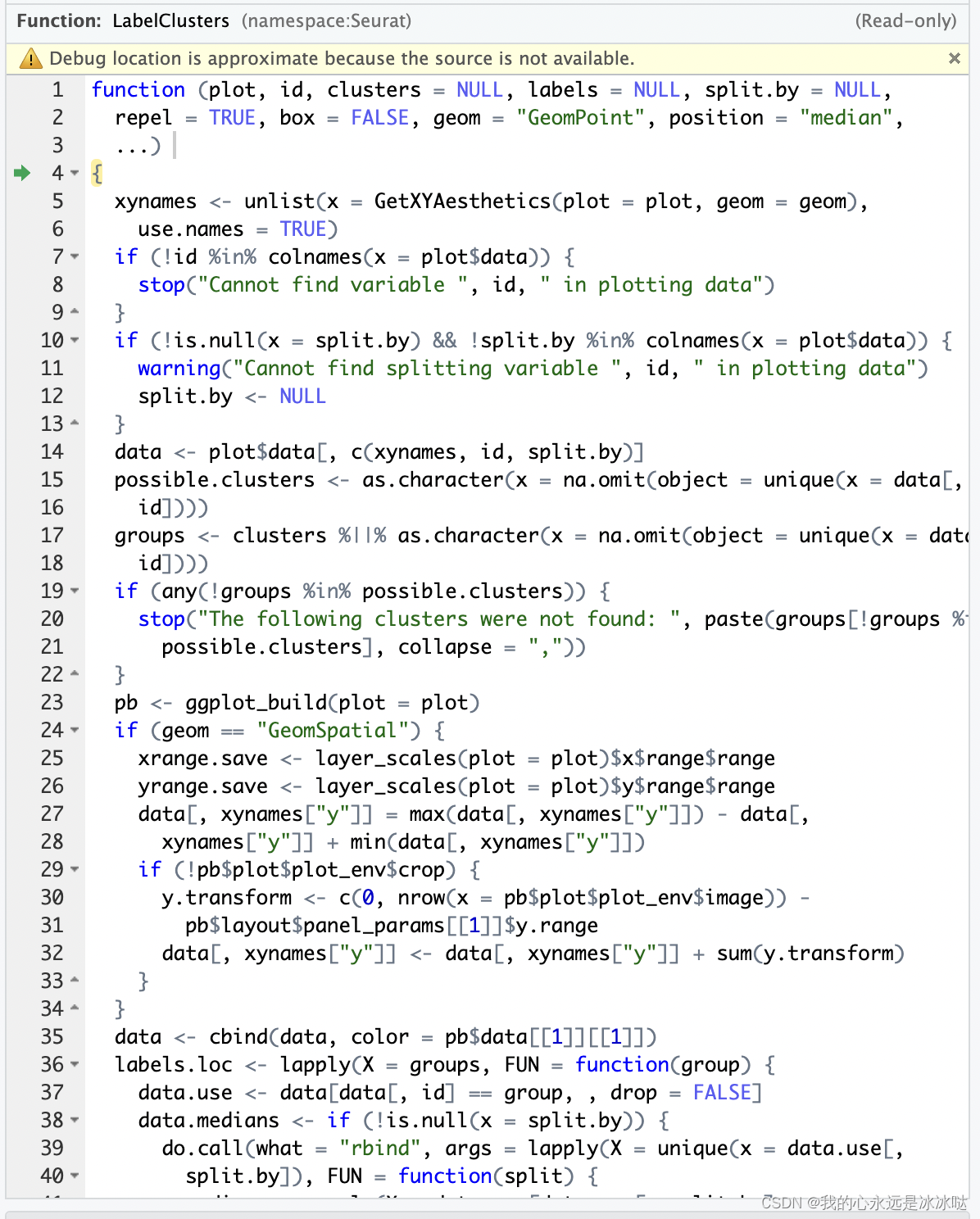
这个函数也是很多条件的,其实核心代码就那么几行, 大致看完这个代码之后,可以着手删代码来
自己实现
将Seurat对象转化到画图的csv文件
write.csv(data,file="./pbmc_UMAP.csv")
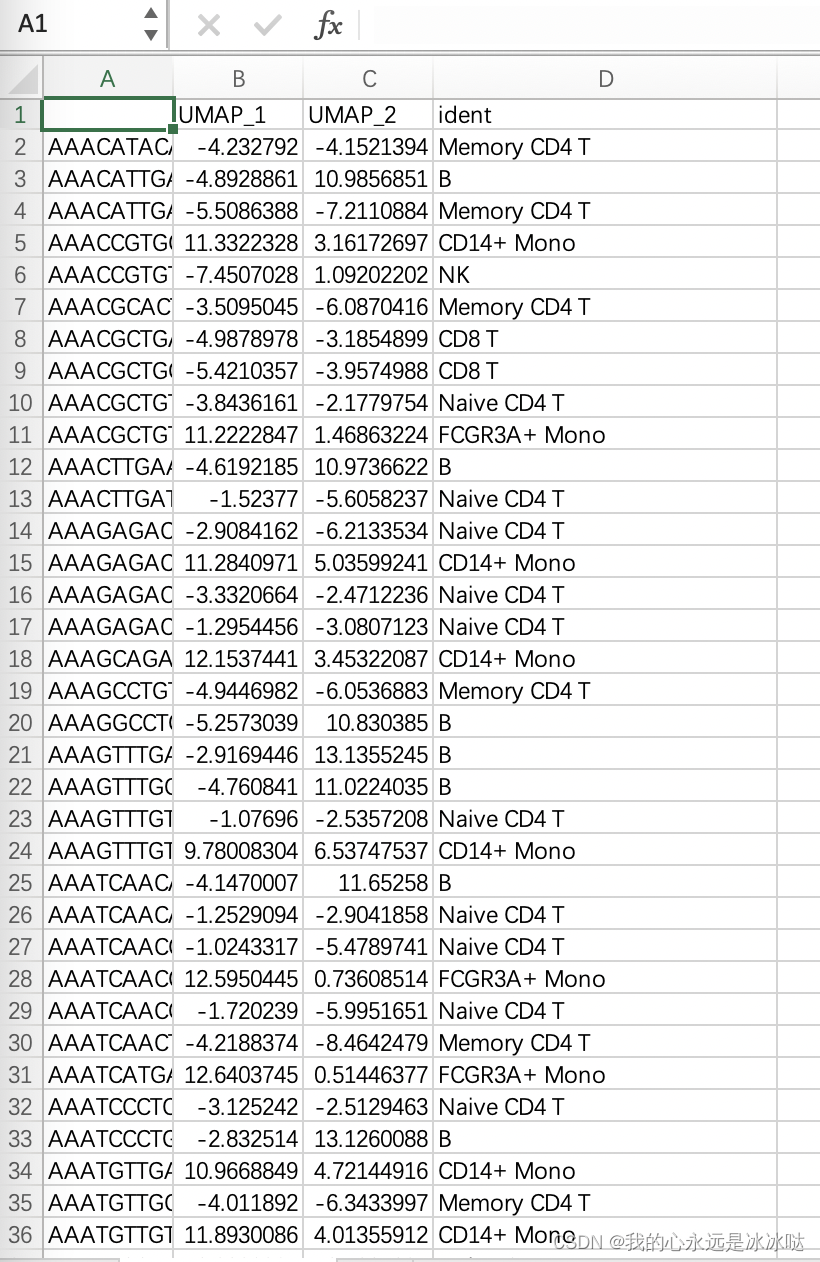
这里其实有一个问题,就是当把Seurat对象存成csv文件,然后再读入时,有些变量的格式是改变的,最常见的就是因子factor类型和chacracter类型,这个一定要注意,否则很容易出错, 比如在Seurat中,data的ident列是因子类型的
 当我把
当我把pbmc_UMAP.csv重新读取进来时,该数据框各列的类型如下
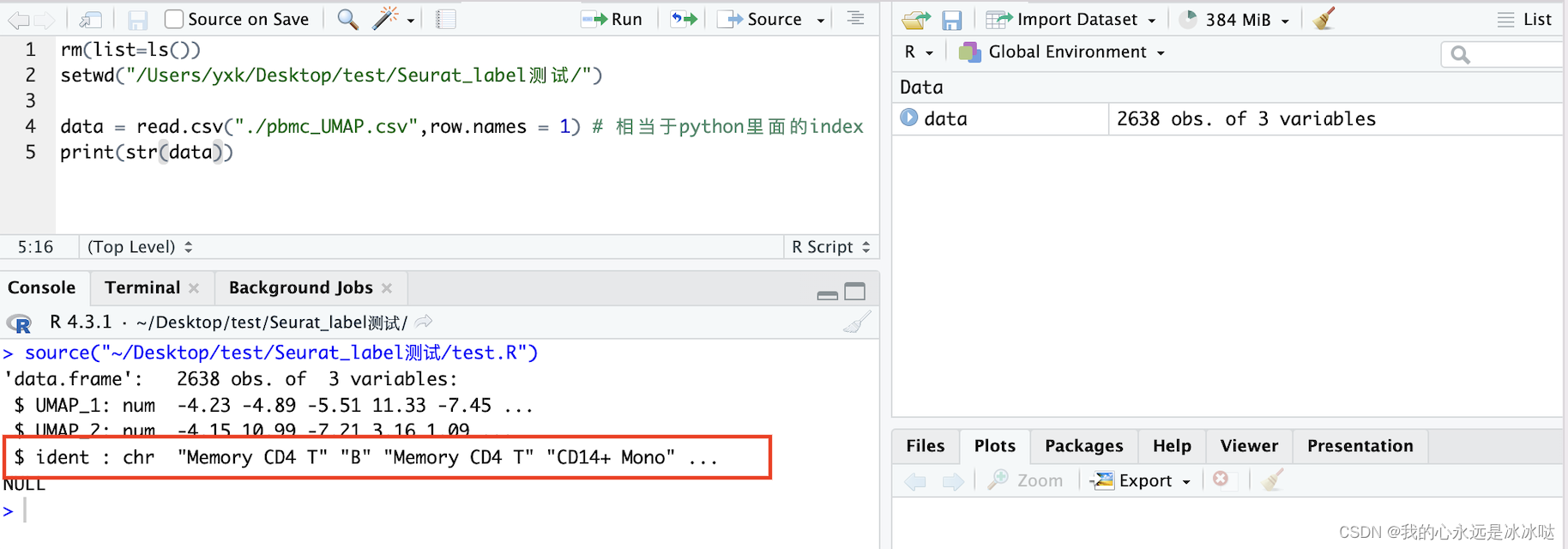 可以看到,我重新读取进来的文件的ident列变成了character类型,这一点一定要多加注意,下面的代码也说明了这一点,需要额外进行类型转换
可以看到,我重新读取进来的文件的ident列变成了character类型,这一点一定要多加注意,下面的代码也说明了这一点,需要额外进行类型转换
简化代码1
rm(list=ls())
setwd("/Users/yxk/Desktop/test/Seurat_label测试/")
suppressPackageStartupMessages(library(ggplot2))
suppressPackageStartupMessages(library(cowplot))
suppressPackageStartupMessages(library(ggrepel))
data = read.csv("./pbmc_UMAP.csv",row.names = 1) # 相当于python里面的index
plot <- ggplot(data = data)
col.by ="ident"
plot = plot + geom_point(mapping = aes_string(x = "UMAP_1",
y = "UMAP_2", color = paste0("`", col.by, "`"), shape = NULL,
alpha = NULL), size = 0.6000758)
plot <- plot + guides(color = guide_legend(override.aes = list(size = 3))) +
labs(color = NULL, title = col.by)
plot <- plot + theme_cowplot()
print(plot)
# clusters这个是有问题的
LabelClusters = function(plot, id, clusters = NULL, labels = NULL, split.by = NULL,
repel = TRUE, box = FALSE, geom = "GeomPoint", position = "median",
...)
{
# xynames <- unlist(x = GetXYAesthetics(plot = plot, geom = geom),
# use.names = TRUE)
xynames=c("UMAP_1","UMAP_2")
names(xynames)=c("x","y")
if (!id %in% colnames(x = plot$data)) {
stop("Cannot find variable ", id, " in plotting data")
}
if (!is.null(x = split.by) && !split.by %in% colnames(x = plot$data)) {
warning("Cannot find splitting variable ", id, " in plotting data")
split.by <- NULL
}
data <- plot$data[, c(xynames, id, split.by)]
possible.clusters <- as.character(x = na.omit(object = unique(x = data[,
id])))
groups <- possible.clusters
if (any(!groups %in% possible.clusters)) {
stop("The following clusters were not found: ", paste(groups[!groups %in%
possible.clusters], collapse = ","))
}
pb <- ggplot_build(plot = plot)
if (geom == "GeomSpatial") {
xrange.save <- layer_scales(plot = plot)$x$range$range
yrange.save <- layer_scales(plot = plot)$y$range$range
data[, xynames["y"]] = max(data[, xynames["y"]]) - data[,
xynames["y"]] + min(data[, xynames["y"]])
if (!pb$plot$plot_env$crop) {
y.transform <- c(0, nrow(x = pb$plot$plot_env$image)) -
pb$layout$panel_params[[1]]$y.range
data[, xynames["y"]] <- data[, xynames["y"]] + sum(y.transform)
}
}
data <- cbind(data, color = pb$data[[1]][[1]])
labels.loc <- lapply(X = groups, FUN = function(group) {
data.use <- data[data[, id] == group, , drop = FALSE]
data.medians <- if (!is.null(x = split.by)) {
do.call(what = "rbind", args = lapply(X = unique(x = data.use[,
split.by]), FUN = function(split) {
medians <- apply(X = data.use[data.use[, split.by] ==
split, xynames, drop = FALSE], MARGIN = 2,
FUN = median, na.rm = TRUE)
medians <- as.data.frame(x = t(x = medians))
medians[, split.by] <- split
return(medians)
}))
}
else {
as.data.frame(x = t(x = apply(X = data.use[, xynames,
drop = FALSE], MARGIN = 2, FUN = median, na.rm = TRUE)))
}
data.medians[, id] <- group
data.medians$color <- data.use$color[1]
return(data.medians)
})
if (position == "nearest") {
labels.loc <- lapply(X = labels.loc, FUN = function(x) {
group.data <- data[as.character(x = data[, id]) ==
as.character(x[3]), ]
nearest.point <- nn2(data = group.data[, 1:2], query = as.matrix(x = x[c(1,
2)]), k = 1)$nn.idx
x[1:2] <- group.data[nearest.point, 1:2]
return(x)
})
}
labels.loc <- do.call(what = "rbind", args = labels.loc)
# labels.loc[, id] <- factor(x = labels.loc[, id], levels = levels(data[,
# id]))
labels.loc$'ident'= as.factor(labels.loc$'ident')
labels <- groups
if (length(x = unique(x = labels.loc[, id])) != length(x = labels)) {
stop("Length of labels (", length(x = labels), ") must be equal to the number of clusters being labeled (",
length(x = labels.loc), ").")
}
names(x = labels) <- groups
for (group in groups) {
labels.loc[labels.loc[, id] == group, id] <- labels[group]
}
#print(labels.loc)
if (box) {
geom.use <- ifelse(test = repel, yes = geom_label_repel,
no = geom_label)
plot <- plot + geom.use(data = labels.loc, mapping = aes_string(x = xynames["x"],
y = xynames["y"], label = id, fill = id), show.legend = FALSE,
...) + scale_fill_manual(values = labels.loc$color[order(labels.loc[,
id])])
}
else {
geom.use <- ifelse(test = repel, yes = geom_text_repel,
no = geom_text)
plot <- plot + geom.use(data = labels.loc, mapping = aes_string(x = xynames["x"],
y = xynames["y"], label = id), show.legend = FALSE,
...)
}
if (geom == "GeomSpatial") {
plot <- suppressMessages(expr = plot + coord_fixed(xlim = xrange.save,
ylim = yrange.save))
}
#print(plot)
return(plot)
}
plot <- LabelClusters(plot = plot, id = "ident", repel = TRUE,
size = 4, split.by = NULL, box = FALSE,
color = "black")
print(plot)
结果如下
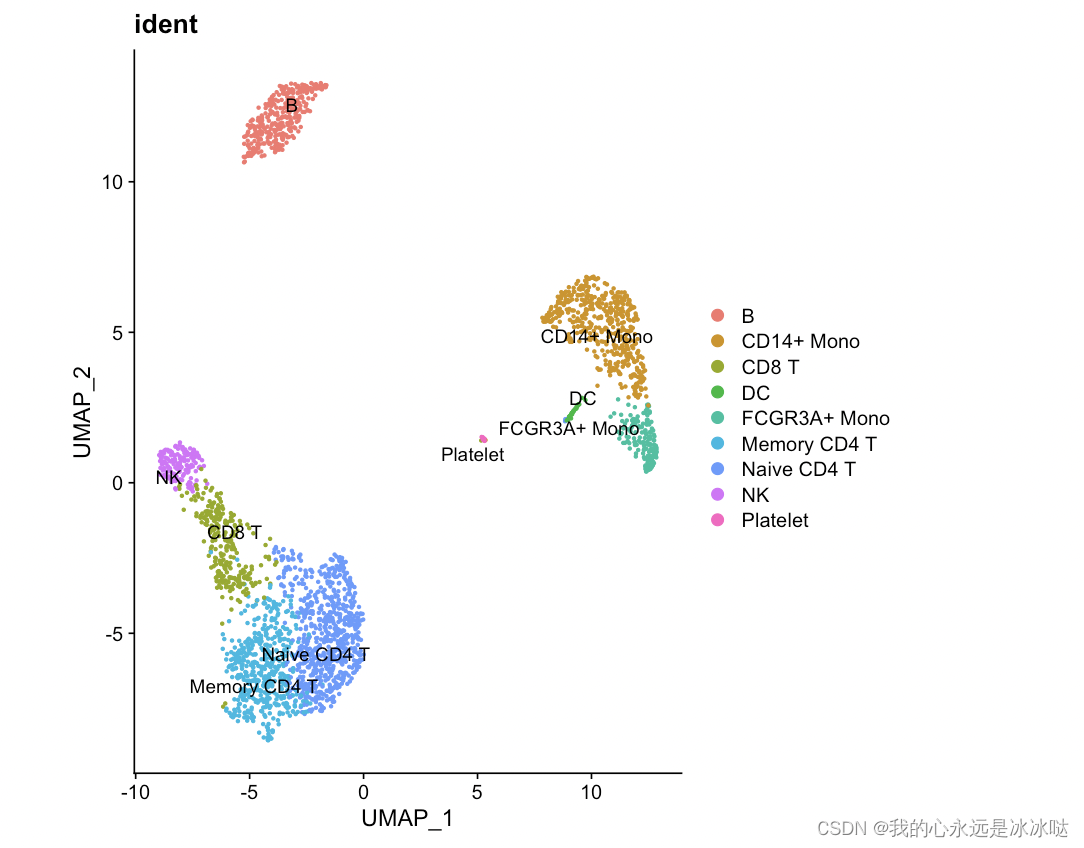
简化代码2
rm(list=ls())
setwd("/Users/yxk/Desktop/test/Seurat_label测试/")
suppressPackageStartupMessages(library(ggplot2))
suppressPackageStartupMessages(library(cowplot))
suppressPackageStartupMessages(library(ggrepel))
data = read.csv("./pbmc_UMAP.csv",row.names = 1) # 相当于python里面的index
plot <- ggplot(data = data)
col.by ="ident"
plot = plot + geom_point(mapping = aes_string(x = "UMAP_1",
y = "UMAP_2", color = paste0("`", col.by, "`"), shape = NULL,
alpha = NULL), size = 0.6000758)
plot <- plot + guides(color = guide_legend(override.aes = list(size = 3))) +
labs(color = NULL, title = col.by)
plot <- plot + theme_cowplot()
print(plot)
LabelClusters = function(plot, id, clusters = NULL, labels = NULL, split.by = NULL,
repel = TRUE, box = FALSE, geom = "GeomPoint", position = "median",
...)
{
# xynames <- unlist(x = GetXYAesthetics(plot = plot, geom = geom),
# use.names = TRUE)
xynames=c("UMAP_1","UMAP_2")
names(xynames)=c("x","y")
data <- plot$data[, c(xynames, id, split.by)]
possible.clusters <- as.character(x = na.omit(object = unique(x = data[, id])))
groups <- possible.clusters
pb <- ggplot_build(plot = plot)
data <- cbind(data, color = pb$data[[1]][[1]])
labels.loc <- lapply(X = groups, FUN = function(group) {
data.use <- data[data[, id] == group, , drop = FALSE]
data.medians <- as.data.frame(x = t(x = apply(X = data.use[, xynames, drop = FALSE], MARGIN = 2, FUN = median, na.rm = TRUE)))
data.medians[, id] <- group
data.medians$color <- data.use$color[1]
return(data.medians)
})
labels.loc <- do.call(what = "rbind", args = labels.loc)
#labels.loc[, id] <- factor(x = labels.loc[, id], levels = levels(data[, id]))
labels.loc$'ident'= as.factor(labels.loc$'ident')
labels <- groups
names(x = labels) <- groups
for (group in groups) {
labels.loc[labels.loc[, id] == group, id] <- labels[group]
}
#print(labels.loc)
geom.use <- ifelse(test = repel, yes = geom_text_repel,
no = geom_text)
plot <- plot + geom.use(data = labels.loc, mapping = aes_string(x = xynames["x"],
y = xynames["y"], label = id), show.legend = FALSE, ...)
return(plot)
}
plot <- LabelClusters(plot = plot, id = "ident", repel = TRUE,
size = 4, split.by = NULL, box = FALSE,
color = "black")
print(plot)
结果如下
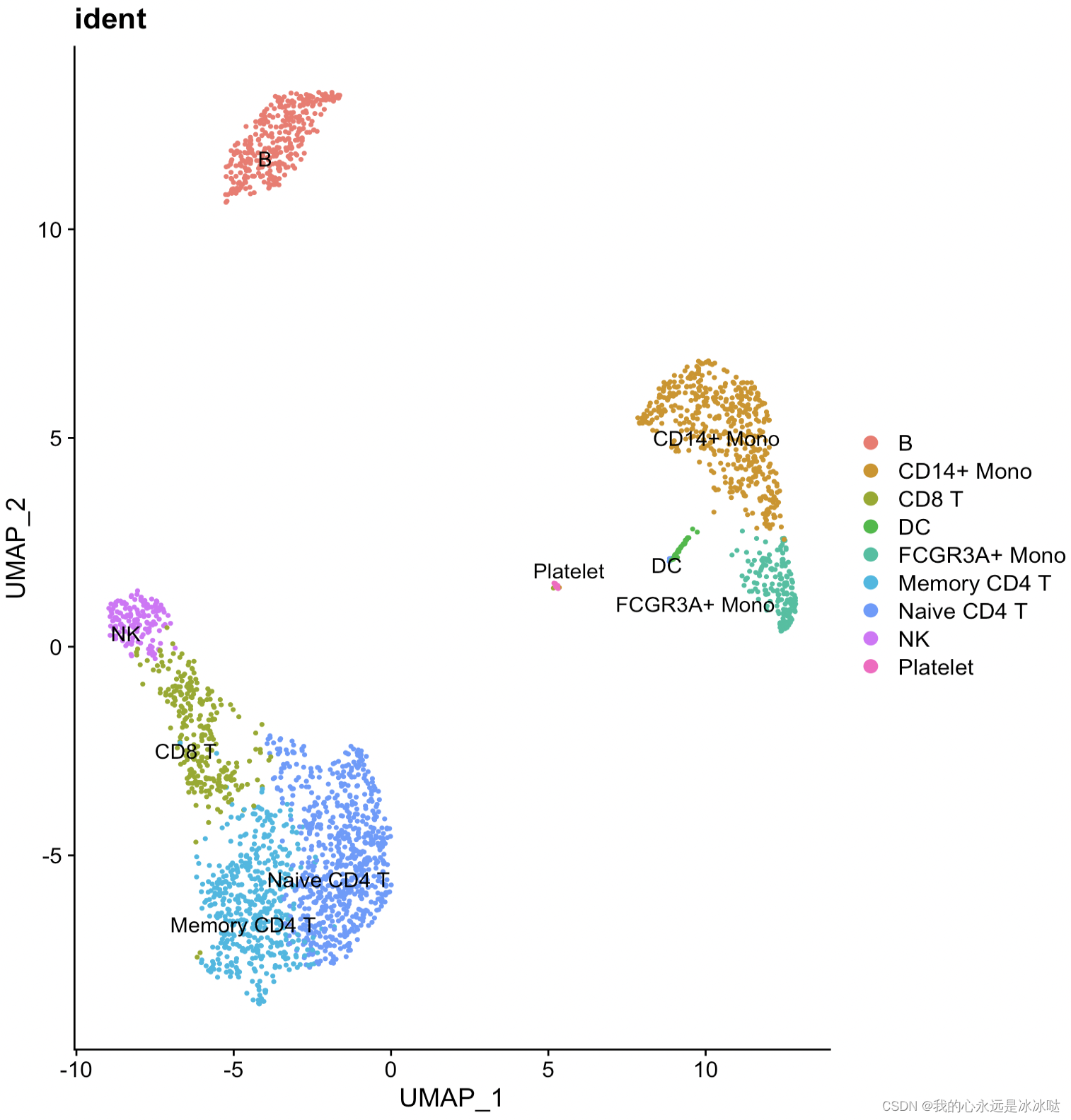
简化代码3
rm(list=ls())
setwd("/Users/yxk/Desktop/test/Seurat_label测试/")
suppressPackageStartupMessages(library(ggplot2))
suppressPackageStartupMessages(library(cowplot))
suppressPackageStartupMessages(library(ggrepel))
data = read.csv("./pbmc_UMAP.csv",row.names = 1) # 相当于python里面的index
plot <- ggplot(data = data)
col.by ="ident"
plot = plot + geom_point(mapping = aes_string(x = "UMAP_1",
y = "UMAP_2", color = paste0("`", col.by, "`"), shape = NULL,
alpha = NULL), size = 0.6000758)
plot <- plot + guides(color = guide_legend(override.aes = list(size = 3))) +
labs(color = NULL, title = col.by)
plot <- plot + theme_cowplot()
print(plot)
###########
id = "ident"
size = 4
color = "black"
xynames=c("UMAP_1","UMAP_2")
names(xynames)=c("x","y")
data <- plot$data[, c(xynames, id)]
clusters<- as.character(x = na.omit(object = unique(x = data[, id])))
groups <- clusters
pb <- ggplot_build(plot = plot)
data <- cbind(data, color = pb$data[[1]][[1]])
labels.loc <- lapply(X = groups, FUN = function(group) {
data.use <- data[data[, id] == group, , drop = FALSE]
data.medians <- as.data.frame(x = t(x = apply(X = data.use[, xynames, drop = FALSE], MARGIN = 2, FUN = median, na.rm = TRUE)))
data.medians[, id] <- group
data.medians$color <- data.use$color[1]
return(data.medians)
})
labels.loc <- do.call(what = "rbind", args = labels.loc)
#labels.loc[, id] <- factor(x = labels.loc[, id], levels = levels(data[, id]))
labels.loc$'ident'= as.factor(labels.loc$'ident')
labels <- groups
names(x = labels) <- groups
for (group in groups) {
labels.loc[labels.loc[, id] == group, id] <- labels[group]
}
#print(labels.loc)
geom.use <- ifelse(test = TRUE, yes = geom_text_repel,
no = geom_text)
plot <- plot + geom.use(data = labels.loc, mapping = aes_string(x = xynames["x"],
y = xynames["y"], label = id), show.legend = FALSE)
print(plot)
结果如下
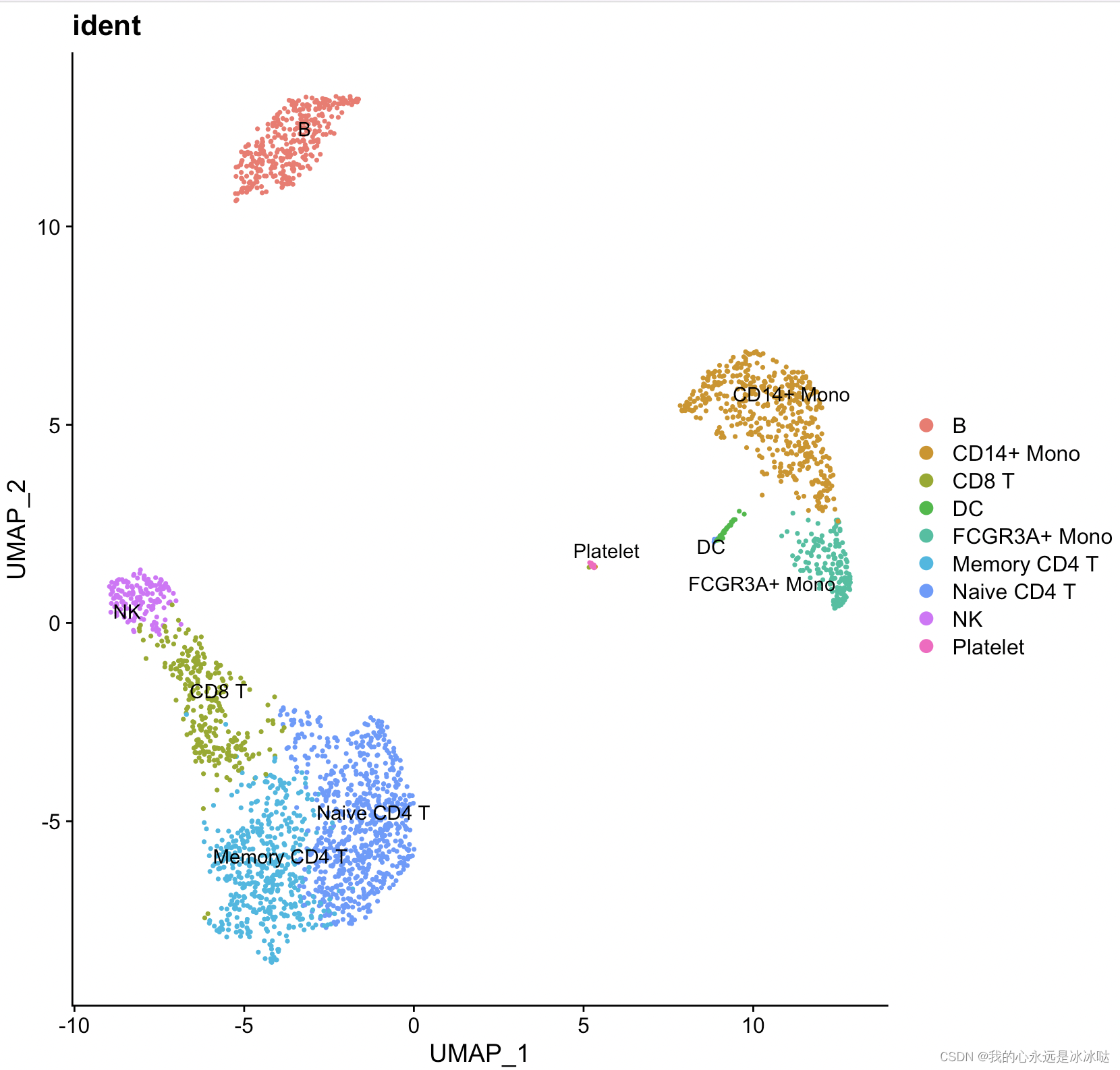


![[OpenGL] opengl切线空间](https://img-blog.csdnimg.cn/direct/d9f9b4cdabc44eb98c3f14a8a93b68e5.png)